Orchid's team of genetic experts has developed a genetic risk score (GRS) for breast cancer.
Written by Orchid Team
Orchid has developed advanced genetic risk scores (GRS) for a variety of diseases. Here we present our data on our GRS of breast cancer.
Breast Cancer
Breast cancer is a cancer that forms in the cells of the breasts [1]. Like other types of cancer, it is caused by cells which begin to divide abnormally. There is a substantial genetic component to breast cancer; individuals with a first degree female relative (mother, sister, or daughter) with the disease are twice as likely to be diagnosed than individuals without a family history [2]. The genetics of breast cancer illustrate both monogenic and polygenic components: there are rarer monogenic variants (such as BRCA1/2) which substantially increase breast cancer risk [3, 4] but there is a polygenic component to breast cancer risk as well, which can sometimes confer risk comparable to monogenic variants [5]. These polygenic scores are still useful for further determining risk among BRCA1/2 carriers [6]. The heritability of breast cancer is approximately 31%, based on an analysis of 200,000 twin pairs drawn from the population registries of Denmark, Norway, Sweden, and Finland [7].
Genetic risk score (GRS)
Orchid’s genetic risk score quantifies the degree to which an individual’s genetics increases their likelihood of developing a specific disease. The GRS for breast cancer includes 1,106,006 variants and was developed based on the variants identified in a study that analyzed genomes of 228,951 individuals of European ancestry [8]. The study included 122,977 cases (individuals with breast cancer) and 105,974 healthy controls [8]. The summary statistics from the meta-analysis were then adjusted for linkage disequilibrium using PRScs [9,10].
Limitations of the GRS
5% to 10% of all breast cancer cases are linked to rare genetic variants in the BRCA1 and BRCA2 genes. Because most of these variants are individually very rare, they are not included in genome-wide association studies for breast cancer and thus they are not captured by our genetic risk score. A low breast cancer GRS is therefore no guarantee for not having an elevated genetic risk for breast cancer. However, more than 90% of all breast cancer cases are influenced by genetic risk factors outside of the BRCA1 and BRCA2 genes, which are captured by our breast cancer GRS. Any individual with a family history of early-onset breast cancer, multiple relatives with breast cancer, or has other risk factors [11] should seek guidance from a genetic counselor with expertise in cancer genetics, since they may carry a rare BRC1/2 variant not captured in Orchid’s GRS.

Clinical Impact and Prevalence
Breast cancer affects over 3.8 million Americans, and 12% of women in the US will be diagnosed in their lifetime [1], [12]. It is usually diagnosed between 55 and 64, with the median age of diagnosis being 62 [13] [14]. The first noticeable symptoms of breast cancer are changes to the breast including a breast lump or thickening that feels different from the surrounding tissue, change in the size, shape or appearance of a breast, and peeling, scaling, or flaking of the pigmented area of skin surrounding the nipple (areola) or breast skin [1]. The prognosis of breast cancer depends on the stage the disease has progressed to when it is diagnosed. Mammography screening can help with earlier detection [15], with different professional associations recommending different start dates for screening. Several treatments such as medications (including chemotherapy), surgery, and radiotherapy may be prescribed by an oncologist [1].
Performant breast cancer risk stratification
Validated using a large cohort of real world women with known breast cancer status
Women in the 99th percentile of genetic risk have a 15.05 percent prevalence of breast cancer, compared to the baseline prevalence of 8.42%. This is lower than the lifetime prevalence of breast cancer reported for US women, as a result of the median age of the UK Biobank (58), which means that many women who will eventually develop breast cancer have not yet done so. This biases the prevalence of breast cancer within the UK Biobank downwards.
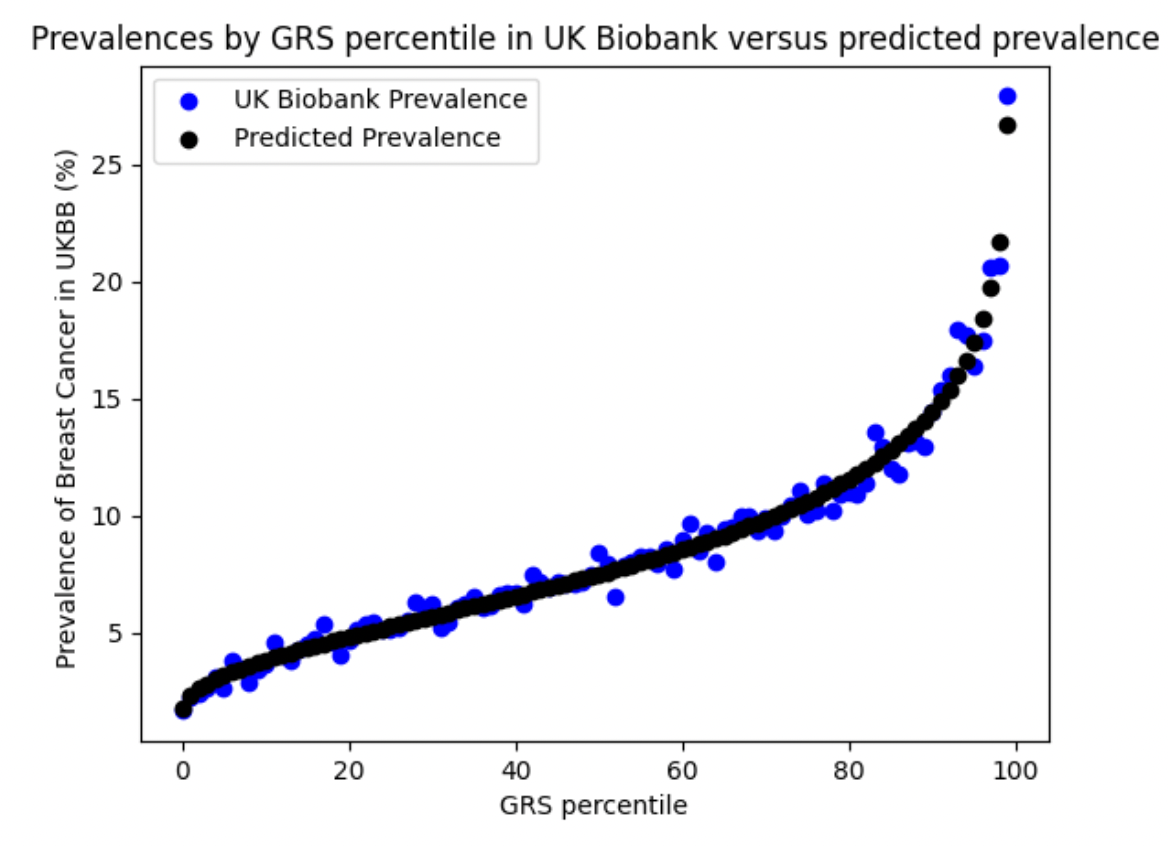
Validation in UK Biobank. In the UK Biobank, cases were identified using self-reported breast cancer (UK Biobank field 20002, code 1002), relevant ICD-9/ICD-10 diagnosis, cancer and death codes. See Supplementary Table for more details. In the validation, prevalence of breast cancer increased with GRS. We restricted our analysis to self-reported British white women whose genetic ancestry matched their self-identification. With our phenotype definition there were 18,588 cases of breast cancer and 202,267 controls.
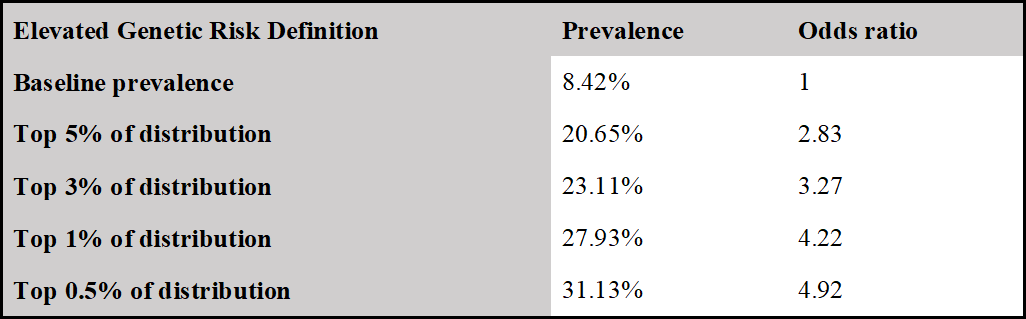
Identification of women at 3 times the baseline risk of breast cancer
Women in the 97th percentile of genetic risk develop breast cancer at 3.32 times the baseline rate. The odds ratio for women in the 99th percentile was 4.22. Baseline rate is the prevalence of the disease in the entire reference population.
Comparison to Published Benchmarks
Orchid’s model achieves greater stratification performance with an AUC of 0.670 compared to the benchmark of 0.644.
We compared the performance of our model as validated on the UK Biobank with the performance of the best model in Khera et al. To make a comparison of models, we restricted our validation sample to those in Phase II of the UK Biobank release, as in Khera et. al. In the first column, we give the results for our predictor with the phenotype as described above. In the second, we report the metrics for the best-performing predictor in Khera et. al using the same phenotype as ours.

1 Khera et al [8]
2 Odds ratio per std. of GRS, controlling for age and PCs, is 1.765 and 1.545 for Orchid and Khera et al. respectively.
Citations
1. Breast cancer. [cited 4 Jan 2022]. Available: https://www.mayoclinic.org/diseases-conditions/breast-cancer/symptoms-causes/syc-20352470
2. CDC. What Are the Risk Factors for Breast Cancer? 21 Sep 2021 [cited 4 Jan 2022]. Available: https://www.cdc.gov/cancer/breast/basic_info/risk_factors.htm
3. Fackenthal JD, Olopade OI. Breast cancer risk associated with BRCA1 and BRCA2 in diverse populations. Nat Rev Cancer. 2007;7: 937–948. doi:10.1038/nrc2054
4. Kuchenbaecker KB, Hopper JL, Barnes DR, Phillips K-A, Mooij TM, Roos-Blom M-J, et al. Risks of Breast, Ovarian, and Contralateral Breast Cancer for BRCA1 and BRCA2 Mutation Carriers. JAMA. 2017;317: 2402–2416. doi:10.1001/jama.2017.7112
5. Mavaddat N, Michailidou K, Dennis J, Lush M, Fachal L, Lee A, et al. Polygenic Risk Scores for Prediction of Breast Cancer and Breast Cancer Subtypes. Am J Hum Genet. 2019;104: 21–34. doi:10.1016/j.ajhg.2018.11.002
6. Barnes DR, Rookus MA, McGuffog L, Leslie G, Mooij TM, Dennis J, et al. Polygenic risk scores and breast and epithelial ovarian cancer risks for carriers of BRCA1 and BRCA2 pathogenic variants. Genet Med. 2020;22: 1653–1666. doi:10.1038/s41436-020-0862-x
7. Mucci LA, Hjelmborg JB, Harris JR, Czene K, Havelick DJ, Scheike T, et al. Familial Risk and Heritability of Cancer Among Twins in Nordic Countries. JAMA. 2016;315: 68–76. doi:10.1001/jama.2015.17703
8. Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50: 1219–1224. doi:10.1038/s41588-018-0183-z
9. Faucon A, Samaroo J, Ge T, Davis LK, Tao R, Cox NJ, et al. Improving the computation efficiency of polygenic risk score modeling: Faster in Julia. bioRxiv. 2021. p. 2021.12.27.474263. doi:10.1101/2021.12.27.474263
10. Ge T, Chen C-Y, Ni Y, Feng Y-CA, Smoller JW. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat Commun. 2019;10: 1776. doi:10.1038/s41467-019-09718-5
11. US Preventive Services Task Force, Owens DK, Davidson KW, Krist AH, Barry MJ, Cabana M, et al. Risk Assessment, Genetic Counseling, and Genetic Testing for BRCA-Related Cancer: US Preventive Services Task Force Recommendation Statement. JAMA. 2019;322: 652–665. doi:10.1001/jama.2019.10987
12. Breastcancer.org. U.S. Breast Cancer Statistics. 4 Feb 2021 [cited 4 Jan 2022]. Available: https://www.breastcancer.org/symptoms/understand_bc/statistics
13. National Cancer Institute. Cancer of the Breast (Female) - Cancer Stat Facts. [cited 4 Jan 2022]. Available: https://seer.cancer.gov/statfacts/html/breast.html
14. American Cancer Society. Breast Cancer Statistics. [cited 4 Jan 2022]. Available: https://www.cancer.org/cancer/breast-cancer/about/how-common-is-breast-cancer.html
15. Siu AL, U.S. Preventive Services Task Force. Screening for Breast Cancer: U.S. Preventive Services Task Force Recommendation Statement. Ann Intern Med. 2016;164: 279–296. doi:10.7326/M15-2886
Appendix: Disease case identification and number of cases in UK Biobank
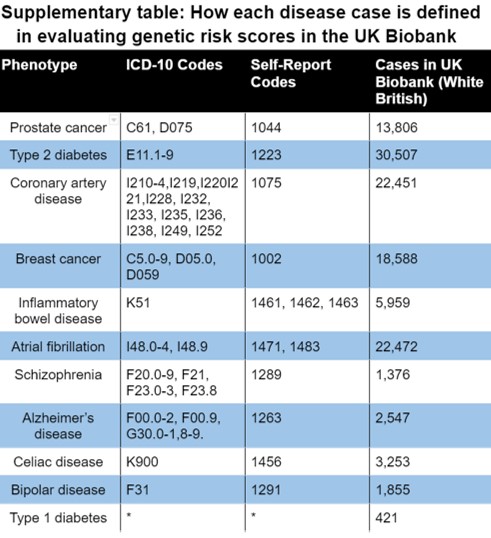
*Type 1 diabetes was defined as a combination the following inclusion and exclusion criteria:
- Self-diagnosed diabetes (any type)
- No self-diagnosed Type 2 diabetes
- Age of diabetes onset between 0 and 20 years
- Started insulin within one year of diagnosis of diabetes







