Orchid's team of genetic experts has developed a genetic risk score (GRS) for childhood asthma.
Written by Orchid Team
Orchid has developed advanced genetic risk scores (GRS) for a variety of diseases. Here we present our data on our GRS of childhood asthma.
Childhood Asthma
Asthma is a common disease of the lungs characterized by inflammation and structural changes to the airways that lead to wheezing, reduced lung function, and heightened sensitivity to allergens. It can develop in either childhood or adulthood, but childhood-onset asthma is more strongly influenced by genetic factors.[1] In addition to genetics, risk factors include prenatal stress and preterm birth, as well as exposure to tobacco smoke both before and after birth.[2]
Genetic Risk Score
Childhood asthma is shaped by both environmental and genetic factors. Monogenic testing is not available because no single gene causes the condition. Genetic risk scores (GRS), which combine the small effects of many variants into a single score, are currently the only way to estimate genetic risk. Although not diagnostic, a GRS can indicate how likely an individual is to develop the disease.
Orchid’s childhood asthma GRS was trained following current industry standards.[3][4] The GRS was constructed using the SBayesRC algorithm trained on publicly available FinnGen and Million Veterans Program summary statistics.[5][6] The summary statistics include 113,450 cases and 813,781 controls.[7] The resulting GRS contains over a million variants.
Risk predictions are adjusted to each individual’s ancestry, with predictive power decaying as genetic distance from the predominately European training data increases.[8] Orchid considers a GRS meaningfully predictive if individuals at roughly the 97.7th percentile have an odds ratio (OR) of 2. The childhood asthma GRS meets this criteria for all common ancestry groups.
Clinical Impact and Prevalence
Asthma affects over 300 million people worldwide and is far more prevalent in high-GDP countries, with prevalence ranging from under 1% in low-GDP regions to around 10% in high-GDP ones. It also much more common near urban centers.[2] In the United States, approximately 6.6% of children under 18 are affected.[9]
Treatments typically include inhaled corticosteroids for long-term control and short-acting bronchodilators for fast-acting symptom reliefs. For children, there is a greater focus on growth monitoring and removal of triggers.[2]
Performant Risk Stratification
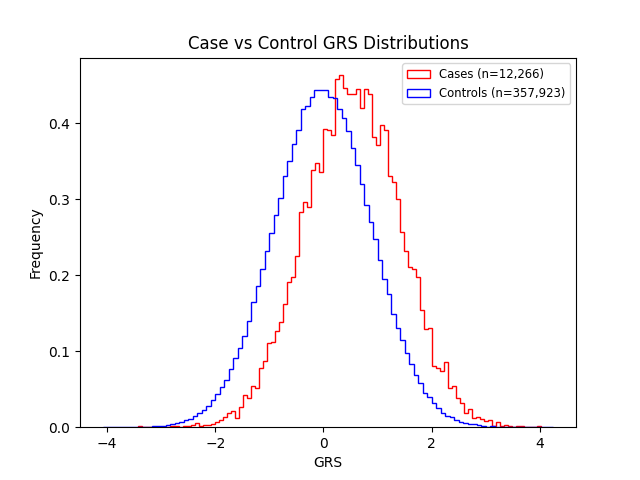
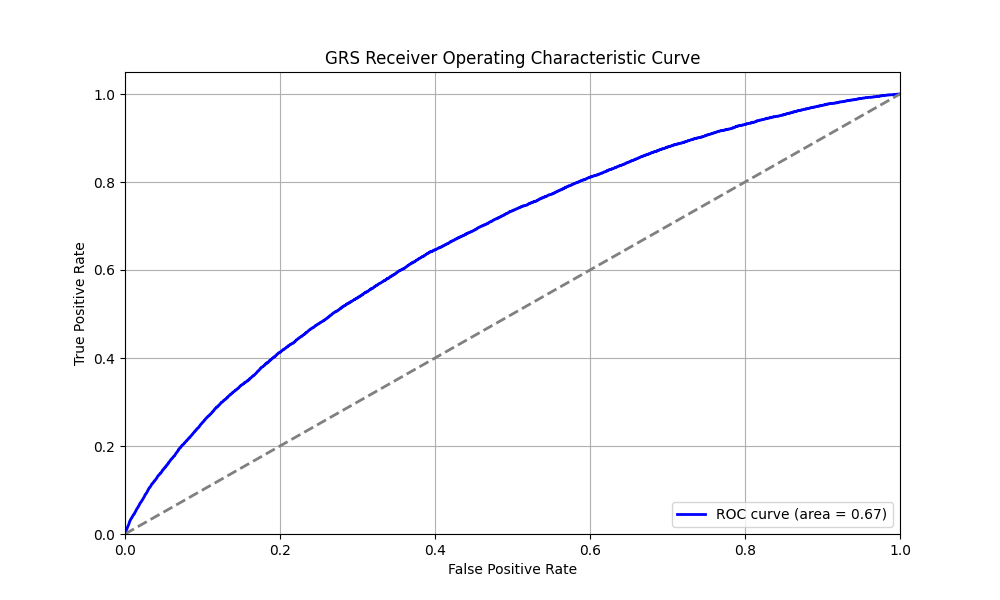
We evaluated the predictive performance of Orchid’s childhood asthma GRS using the UK Biobank (UKB), a research database of roughly 500,000 genotyped individuals from the United Kingdom. We restricted the analysis to participants of British ancestry and excluded those who reported developing asthma after age 18. Cases (12,266) were defined as participants who self-reported asthma onset by age 18, and controls (356,923) as participants who self-reported never having asthma. We then grouped individuals by GRS percentile and compared the observed disease prevalence within each group to our model’s predictions (Figure 1). For additional technical details, see the Supplementary Data.
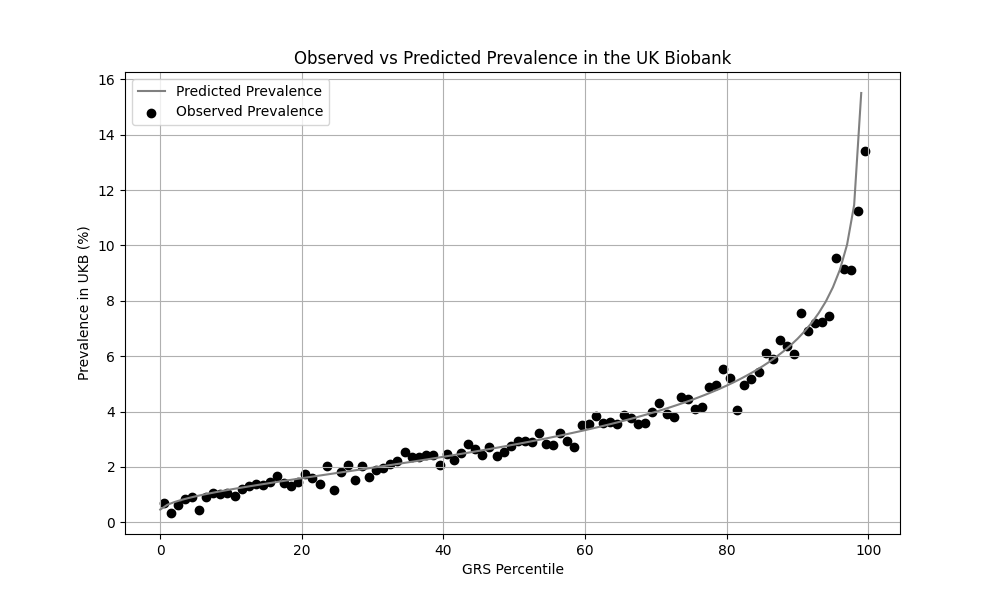
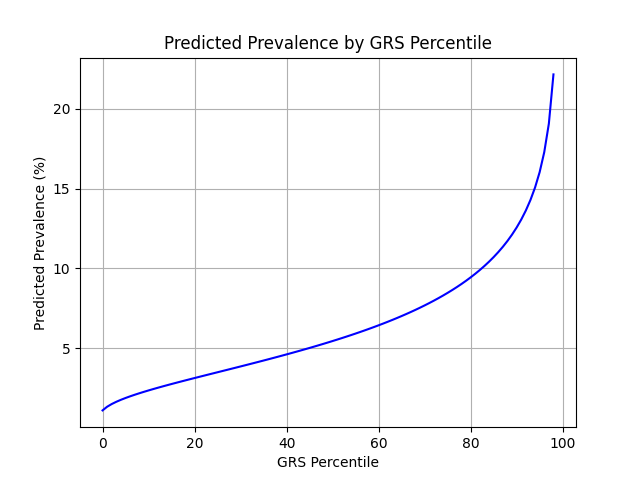
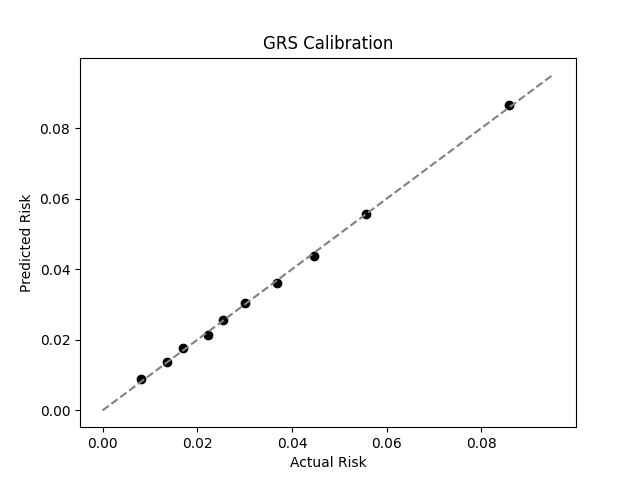
UKB participants tend to be healthier than the general population, which leads to lower observed disease prevalence.[10] The CDC estimates that approximately 6.5% of children have asthma, higher than the prevalence in the UKB.[9] We adjust our model so that its average predicted risk aligns with this estimate (see Figure 2).[11] People at the tail end of the GRS distribution were at an elevated risk compared to the mean (see Table 3), with children in the 99th percentile 3.4x more likely to develop childhood asthma than average (22.1% vs 6.5%).
References
1. Pividori M, Schoettler N, Nicolae DL, et al. Shared and distinct genetic risk factors for childhood-onset and adult-onset asthma. Lancet Respir Med. 2019;7:509–522. doi:10.1016/S2213-2600(19)30055-4.
2. Holgate ST, Wenzel S, Postma DS, et al. Asthma. Nat Rev Dis Primers. 2015;1:15025. doi:10.1038/nrdp.2015.25.
3. Moore S, Davidson I, Anomaly J, et al. Development and validation of polygenic scores for within-family prediction of disease risks. medRxiv. 2025. doi:10.1101/2025.08.06.25333145.
4. Cordogan S, Starr DB, Treff NR, et al. Within- and between-family validation of nine polygenic risk scores developed in 1.5 million individuals: implications for IVF, embryo selection, and reduction in lifetime disease risk. medRxiv. 2025. doi:10.1101/2025.10.24.25338613.
5. Zheng, Z., Liu, S., Sidorenko, J. et al. Leveraging functional genomic annotations and genome coverage to improve polygenic prediction of complex traits within and between ancestries. Nat Genet 56, 767–777 (2024). https://doi.org/10.1038/s41588-024-01704-y
6. FinnGen. FinnGen+MVP+UKBB Summary Statistics. Available at: https://mvp-ukbb.finngen.fi/about. Accessed 2025-12-05.
7. FinnGen. FinnGen+MVP+UKBB Phenotypes. Available at: https://mvp-ukbb.finngen.fi. Accessed 2025-12-05.
8. Privé, Florian et al. “Portability of 245 polygenic scores when derived from the UK Biobank and applied to 9 ancestry groups from the same cohort.” American journal of human genetics vol. 109,1 (2022): 12-23. doi:10.1016/j.ajhg.2021.11.008
9. CDC. Most recent national asthma data. May 10 2023 [cited 15 Dec 2025]. https://www.cdc.gov/asthma/most_recent_national_asthma_data.htm.
10. Fry A, Littlejohns TJ, Sudlow C, et al. Comparison of sociodemographic and health-related characteristics of UK Biobank participants with those of the general population. Am J Epidemiol. 2017;186:1026–1034. doi:10.1093/aje/kwx246.
11. Chatterjee N, Shi J, García-Closas M et al. Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat Rev Genet. 2016;17:392–406. doi:10.1038/nrg.2016.27
Supplementary Figures
Acknowledgements
This research has been conducted using the UK Biobank Resource under Application Number 80545.