Orchid's team of genetic experts has released a genetic risk score (GRS) for multiple sclerosis.
Written by Orchid Team
Orchid has developed advanced genetic risk scores (GRS) for a variety of diseases. Here we present our data on our GRS of multiple sclerosis.
Multiple Sclerosis
Multiple sclerosis (MS) is a disease of the central nervous system. It is often characterized by temporary vision loss, weakness, and sensory/autonomic dysfunction. The initial symptoms typically manifest in patients between 20-40 years old as reversible episodes of neurological dysfunction lasting several days or weeks. Irreversible damage occurs as the disease progresses. MS-related complications account for over half of deaths among patients with MS, with patients living 7-14 fewer years than average. Common risk factors include low vitamin D, smoking, and obesity.[1]
Genetic Risk Score
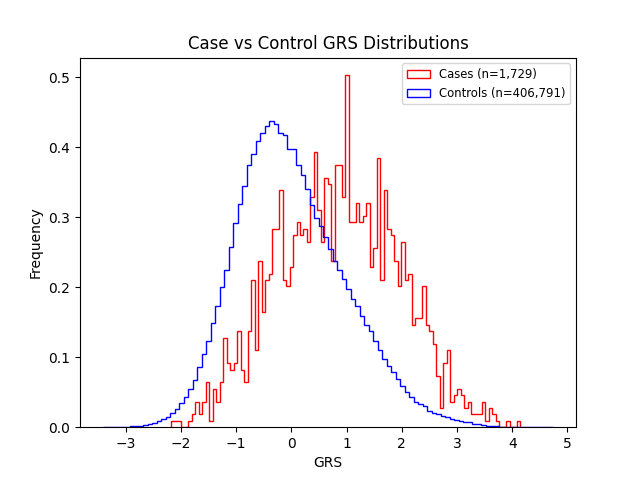
MS is shaped by both environmental and genetic factors. Monogenic testing is not available because no single gene causes the condition. Genetic risk scores (GRS), which combine the small effects of many variants into a single score, are currently the only way to estimate genetic risk. Although not diagnostic, a GRS can indicate how likely an individual is to develop the disease. Orchid’s MS GRS evaluates hundreds of thousands of variants and is available to parents of European ancestry who have at least one first-degree relative diagnosed with MS.
Clinical Impact and Prevalence
MS affects over 2 million people worldwide and approximately 0.3% of adults in the US.[2] While there are a number disease-modifying treatments that may slow down the development of MS, there is no cure. However, there are a number of environmental risk factors that one can minimize, most notably: smoking, adolescent obesity, and vitamin D deficiency. Some of these risk factors have shown gene-environment interactions, suggesting that these interventions may be more effective if one has a high genetic risk.[1]
Performant Multiple Sclerosis Risk Stratification
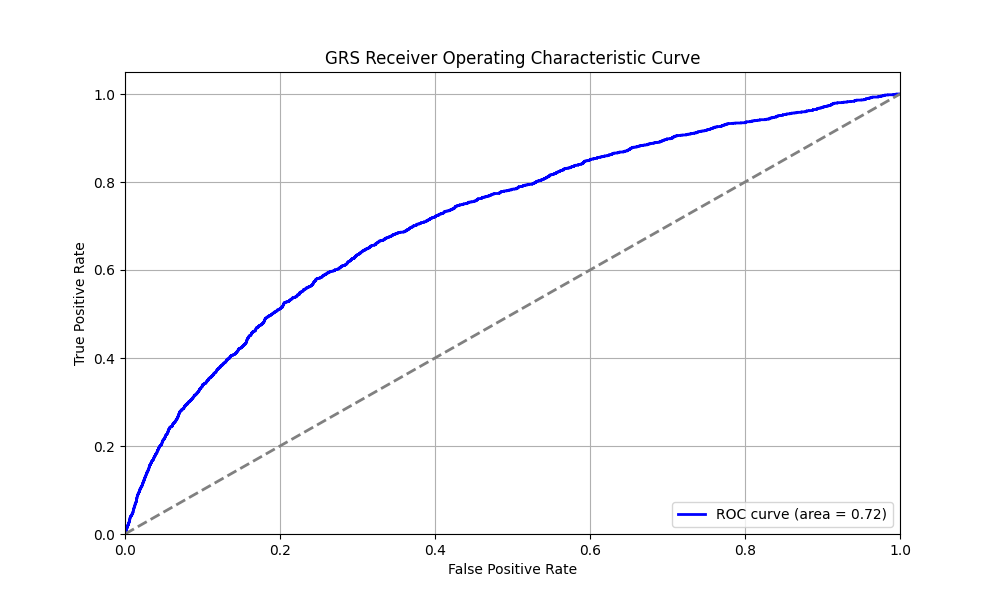
We evaluated the predictive performance of Orchid’s MS GRS using the UK Biobank (UKB), a research database of roughly 500,000 genotyped individuals from the United Kingdom. We restricted the analysis to participants of British ancestry and defined MS using the G35 ICD-10 code, yielding 1,729 cases and 406,791 controls (0.4% prevalence). We then grouped individuals by GRS percentile and compared the observed disease prevalence within each group to our model’s predictions (Figure 1). For additional technical details, see the Supplementary Data.
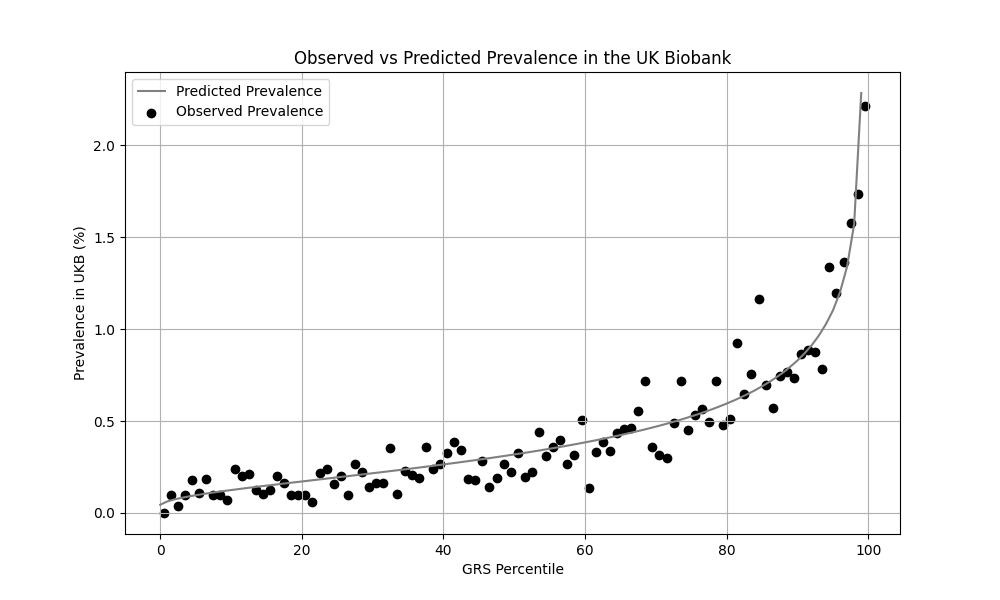
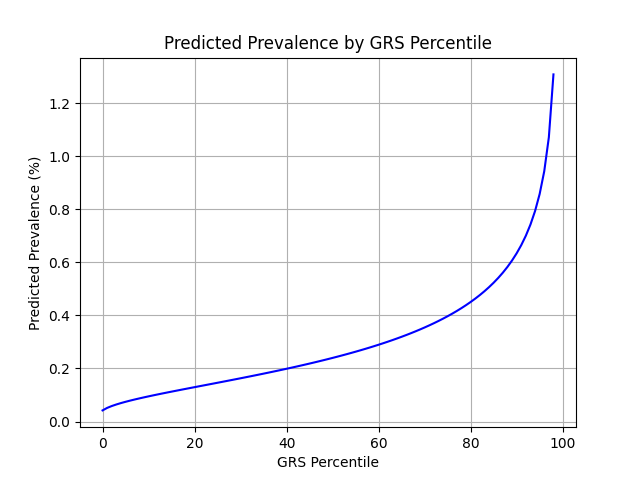
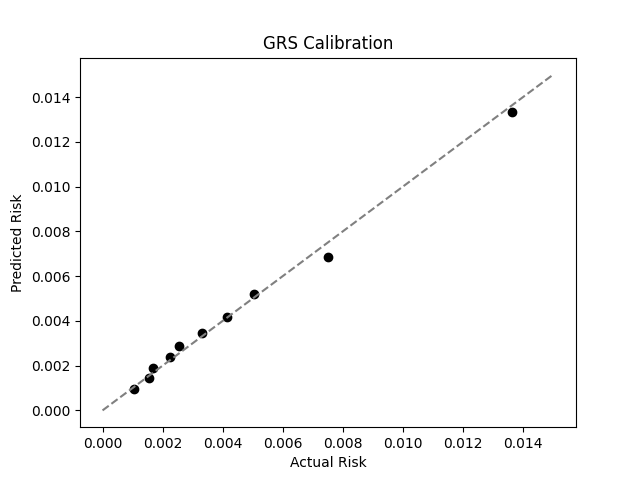
Wallin et al. estimate a 0.3% prevalence of MS in the US,[2] similar to the computed 0.4% prevalence in the UKB. We adjust our model so that its average prevalence aligns with the Wallin et al. estimate (see Figure 2).[3] People at the tail end of the GRS distribution were at an elevated risk compared to the mean (see Table 3), with adults in the 99th percentile 4.2x more likely to develop IBD than average (1.3% vs 0.3%).
References
1. Filippi M, Bar-Or A, Piehl F, et al. Multiple sclerosis. Nat Rev Dis Primers. 2018;4:43. doi:10.1038/s41572-018-0041-4.
2. Wallin MT, Culpepper WJ, Campbell JD, et al. The prevalence of MS in the United States: A population-based estimate using health claims data. Neurology. 2019;92(10):e1029–e1040. doi:10.1212/WNL.0000000000007035.
3. Chatterjee N, Shi J, García-Closas M et al. Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat Rev Genet. 2016;17:392–406. doi:10.1038/nrg.2016.27
Supplementary Data
Acknowledgements
This research has been conducted using the UK Biobank Resource under Application Number 80545.