Orchid's team of genetic experts has developed a genetic risk score (GRS) for Prostate Cancer.
Written by Orchid Team
Orchid has developed advanced genetic risk scores (GRS) for a variety of diseases. Here we present our data on our GRS of Prostate Cancer.
Prostate Cancer
Prostate cancer is a form of cancer that affects the prostate, a gland in the male reproductive system [1]. Like other types of cancer, it is caused by cells which begin to divide abnormally [2]. Genetics are implicated in the development of prostate cancer, the heritability of which is estimated to be between 52% to 63% based on an analysis of almost 50,000 twin pairs drawn from the Nordic Twin Study of Cancer [3].
Genetic risk score (GRS)
A genetic risk score quantifies the degree to which an individual’s genetics increases their likelihood of developing a specific disease. The GRS for prostate cancer includes 1,106,020 variants and was developed by selecting the genome-wide significant SNPs identified in a study that analyzed genomes of about 74,849 individuals of European ancestry [4]. The study included 46,939 cases (individuals with prostate cancer) and 27,910 healthy controls [4]. The summary statistics from the meta-analysis were then adjusted for linkage disequilibrium using PRScs.

Clinical Impact and Prevalence
Prostate cancer affects roughly 3.2 million American men, and 12.5% will be diagnosed at some point during their lifetime [5], making it the most common non-skin cancer in men [6]. The average age of diagnosis is 66 [7]. Typical symptoms of prostate cancer include frequent and difficult urination, pain in and around the groin, and pain in adjacent parts of the body such as the pelvis and hips [8]. Prostate cancer is very common and about 80% of men diagnosed with prostate cancer will live at least five years after their diagnosis [9]. Many cases of prostate cancer do not require active treatment, but some may require medical treatment by an oncologist, surgery, and/or radiotherapy [8].
Performant prostate cancer risk stratification
Validated using a large cohort of real world men with known prostate cancer status
Within the UK Biobank, men in the 99th percentile of genetic risk have a 30.5 percent absolute risk of prostate cancer, compared to a baseline prevalence of 7.4 percent. This is different from the lifetime prevalence figure reported above mainly because the UK Biobank cohort has a median age of 58 [10], so the majority of men who will develop prostate cancer have not yet done so, or it has not been detected yet. This will lower the prevalence of prostate cancer in the UK Biobank cohort.

In the UK Biobank, cases were identified using self-reported prostate cancer (UK Biobank field 20002) relevant ICD-10 diagnosis and death codes. See supplementary table for more details. In the validation, prevalence of the disease increased with GRS. Using our phenotype definition, in our sample of self-reported white British men, there were 13,806 cases of prostate cancer and 173,859 controls (prevalence of 7.36%).
People at the tail end of GRS distribution were at an elevated risk for developing the disease in comparison to the baseline rate. For each GRS threshold, we also computed the odds ratio (with the baseline rate as a comparison group). Baseline rate is the prevalence of the disease in the entire reference population.
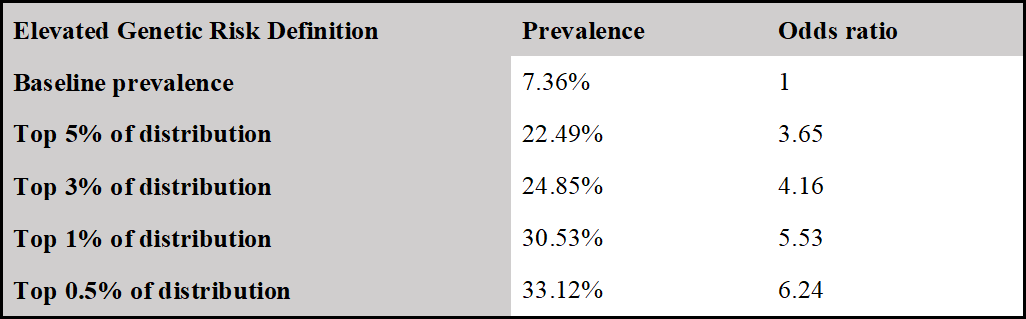
Identification of men at 4 times the baseline risk of prostate cancer
Men in the 99th percentile of genetic risk develop prostate cancer at 4.15 times the baseline rate, with an odds ratio of 5.53. Baseline rate is the prevalence of the disease in the entire reference population.
Comparison to Published Benchmarks
Orchid’s model achieves comparable stratification performance with an AUC of 0.683 compared to the benchmark at 0.662.
We compared the performance of our model as validated on the UK Biobank with the performance of the best model in Jia et al [11]. To make a comparison of models, we restricted our validation sample to those in Phase II of the UK Biobank release, as in Jia et al. In the first column, we give the results for our predictor with the phenotype as described above. In the second, we report the metrics for the best-performing predictor in Jia et al. from their paper.
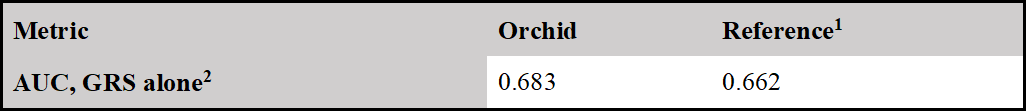
1 Jia et al [11]
2 Jia et al (Jia et al. 2020) did not provide odds per std. of GRS, so cannot make a direct comparison.
Citations
1. CDC. What Is Prostate Cancer? 23 Aug 2021 [cited 4 Jan 2022]. Available: https://www.cdc.gov/cancer/prostate/basic_info/what-is-prostate-cancer.htm
2. CDC. What Causes Prostate Cancer? [cited 4 Jan 2022]. Available: https://www.cancer.org/cancer/prostate-cancer/causes-risks-prevention/what-causes.html
3. Hjelmborg JB, Scheike T, Holst K, Skytthe A, Penney KL, Graff RE, et al. The heritability of prostate cancer in the Nordic Twin Study of Cancer. Cancer Epidemiol Biomarkers Prev. 2014;23. doi:10.1158/1055-9965.EPI-13-0568
4. Schumacher FR, Al Olama AA, Berndt SI, Benlloch S, Ahmed M, Saunders EJ, et al. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat Genet. 2018;50: 928–936.
5. National Cancer Institute. Cancer of the Prostate - Cancer Stat Facts. [cited 4 Jan 2022]. Available: https://seer.cancer.gov/statfacts/html/prost.html
6. Nelen V. Epidemiology of prostate cancer. Recent Results Cancer Res. 2007;175: 1–8.
7. American Cancer Society. Key Statistics for Prostate Cancer. [cited 4 Jan 2022]. Available: https://www.cancer.org/cancer/prostate-cancer/about/key-statistics.html
8. CDC. How Is Prostate Cancer Treated? 13 Sep 2021 [cited 4 Jan 2022]. Available: https://www.cdc.gov/cancer/prostate/basic_info/treatment.htm
9. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68: 7–30.
10. : Data-Field 21022. [cited 22 Feb 2022]. Available: https://biobank.ndph.ox.ac.uk/ukb/field.cgi?id=21022
11. Jia G, Lu Y, Wen W, Long J, Liu Y, Tao R, et al. Evaluating the Utility of Polygenic Risk Scores in Identifying High-Risk Individuals for Eight Common Cancers. JNCI Cancer Spectr. 2020;4: kaa021.
Appendix: Disease case identification and number of cases in UK Biobank
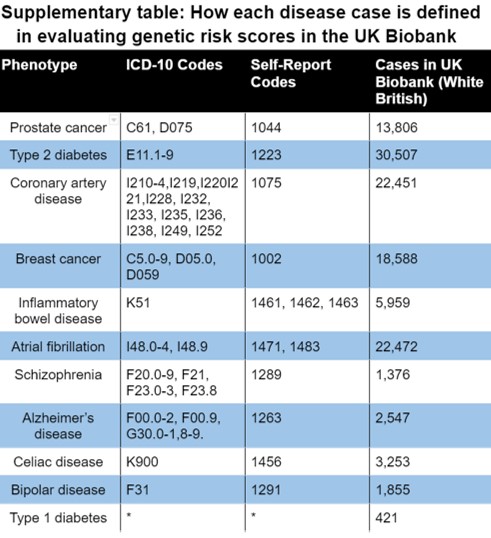
*Type 1 diabetes was defined as a combination the following inclusion and exclusion criteria:
- Self-diagnosed diabetes (any type)
- No self-diagnosed Type 2 diabetes
- Age of diabetes onset between 0 and 20 years
- Started insulin within one year of diagnosis of diabetes







